- Learning to Segment Fine Structures Under Image-Level Supervision With an Application to Nematode Segmentation
Long Chen, Martin Strauch, Matthias Daub, Hans-Georg Luigs, Marcus Jansen and Dorit Merhof
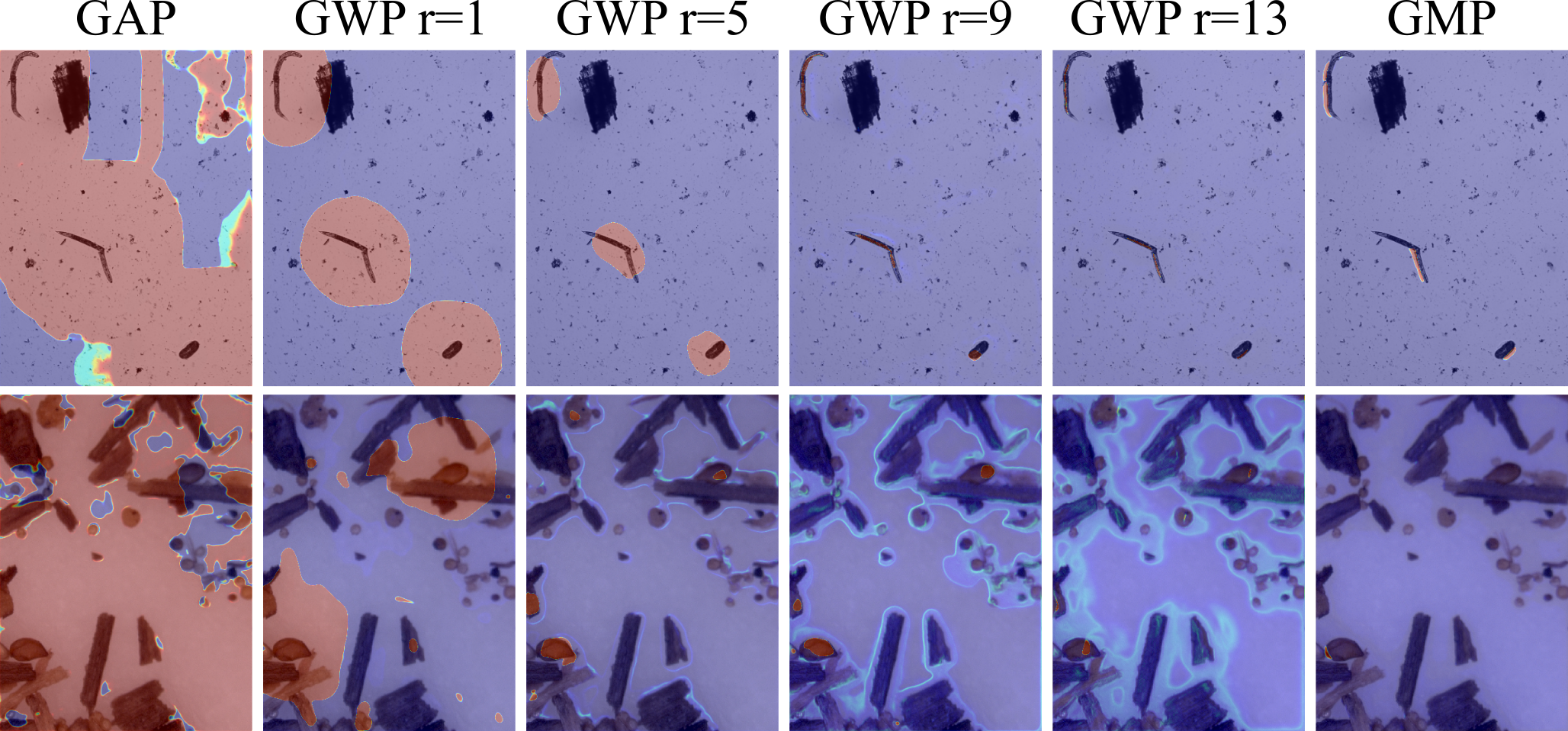
Abstract: Image segmentation models trained only with image-level labels have become increasingly popular as they require significantly less annotation effort than models trained with scribble, bounding box or pixel-wise annotations. While methods utilizing image-level labels achieve promising performance for the segmentation of larger-scale objects, they perform less well for the fine structures frequently encountered in biological images. In order to address this performance gap, we propose a deep network architecture based on two key principles, Global Weighted Pooling (GWP) and segmentation refinement by low-level image cues, that, together, make segmentation of fine structures possible. We apply our segmentation method to image datasets containing such fine structures, nematodes (worms + eggs) and nematode cysts immersed in organic debris objects, which is an application scenario encountered in automated soil sample screening. Supervised only with imagelevel labels, our approach achieves Dice coefficients of 79.72% and 58.51% for nematode and nematode cyst segmentation, respectively.
- Semi-supervised Instance Segmentation with a Learned Shape Prior
Long Chen, Weiwen Zhang, Yuli Wu, Martin Strauch and Dorit Merhof
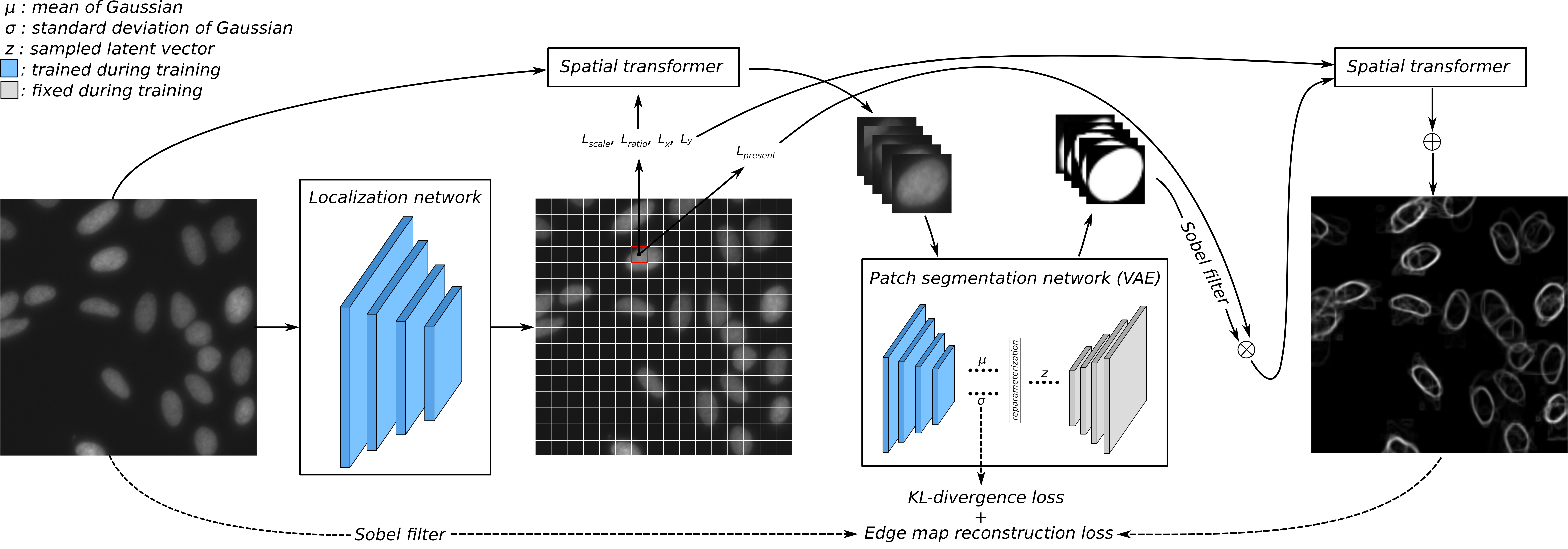
Abstract: To date, most instance segmentation approaches are based on supervised learning that requires a considerable amount of annotated object contours as training ground truth. Here, we propose a framework that searches for the target object based on a shape prior. The shape prior model is learned with a variational autoencoder that requires only a very limited amount of training data: In our experiments, a few dozens of object shape patches from the target dataset, as well as purely synthetic shapes, were suffcient to achieve results en par with supervised methods with full access to training data on two out of three cell segmentation datasets. Our method with a synthetic shape prior was superior to pre-trained supervised models with access to limited domain-speci c training data on all three datasets. Since the learning of prior models requires shape patches, whether real or synthetic data, we call this framework semi-supervised learning.
- Improving Pixel Embedding Learning through Intermediate Distance Regression Supervision
Yuli Wu, Long Chen and Dorit Merhof
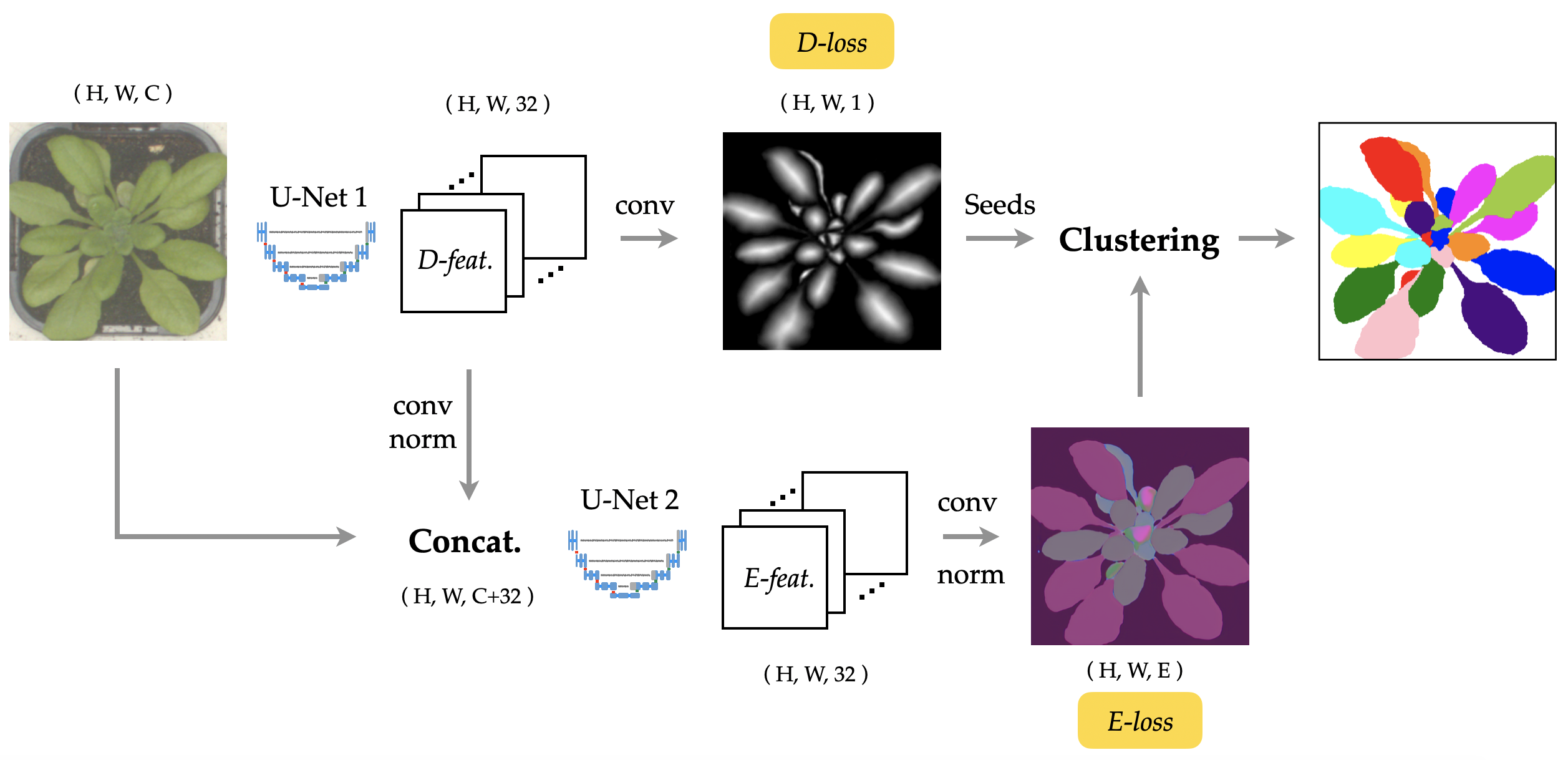
Abstract: As a proposal-free approach, instance segmentation through pixel embedding learning and clustering is gaining more emphasis. Compared with bounding box re nement approaches, such as Mask R-CNN, it has potential advantages in handling complex shapes and dense objects. In this work, we propose a simple, yet highly effective, architecture for object-aware embedding learning. A distance regression module is incorporated into our architecture to generate seeds for fast clustering. At the same time, we show that the features learned by the distance regression module are able to promote the accuracy of learned object- aware embeddings signi cantly. By simply concatenating features of the distance regression module to the images as inputs of the embedding module, the mSBD scores on the CVPPP Leaf Segmentation Challenge can be further improved by more than 8% compared to the identical setup without concatenation, yielding the best overall result amongst the leaderboard at CodaLab.
- A CNN Framework Based on Line Annotations for Detecting Nematodes in Microscopic Images
Long Chen, Martin Strauch, Matthias Daub, Xiaochen Jiang, Marcus Jansen, Hans-Georg Luigs, Susanne Schultz-Kuhlmann, Stefan Kruessel and Dorit Merhof
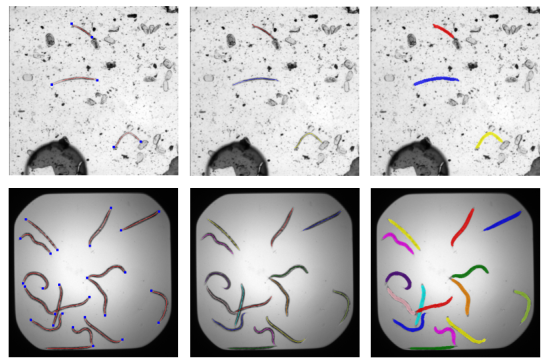
Abstract: Plant parasitic nematodes cause damage to crop plants on a global scale. Robust detection on image data is a prerequisite for monitoring such nematodes, as well as for many biological studies involving the nematode C. elegans, a common model organism. Here, we propose a framework for detecting worm-shaped objects in microscopic images that is based on convolutional neural networks (CNNs). We annotate nematodes with curved lines along the body, which is more suitable for worm-shaped objects than bounding boxes. The trained model predicts worm skeletons and body endpoints. The endpoints serve to untangle the skeletons from which segmentation masks are reconstructed by estimating the body width at each location along the skeleton. With light-weight backbone networks, we achieve 75.85% precision, 73.02% recall on a potato cyst nematode data set and 84.20% precision, 85.63% recall on a public C. elegans data set.