- Instance Segmentation of Biomedical Images with an Object-aware Embedding Learned with Local Constraints
Long Chen, Martin Strauch and Dorit Merhof
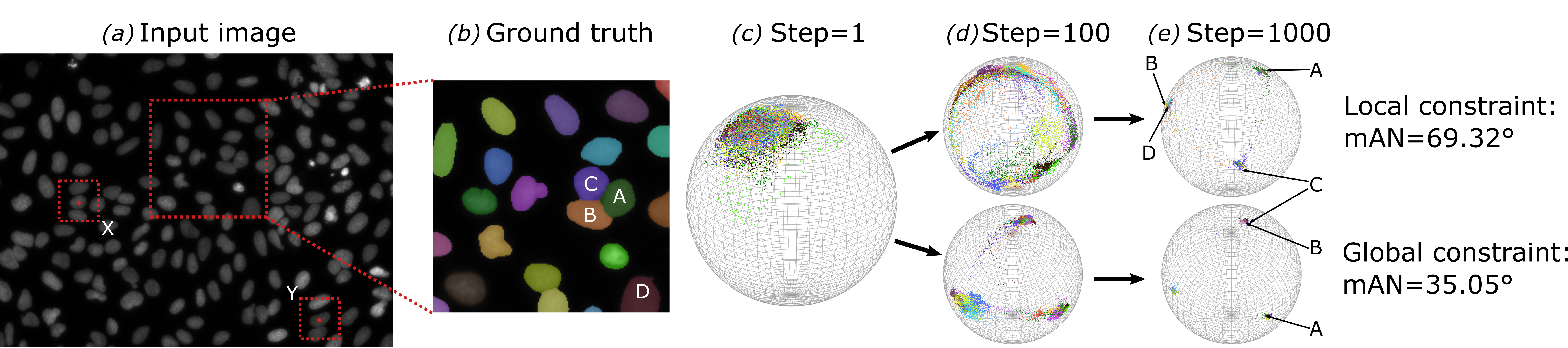
Abstract: Automatic instance segmentation is a problem that occurs in many biomedical applications. State-of-the-art approaches either perform semantic segmentation or re ne object bounding boxes obtained from detection methods. Both su er from crowded objects to varying degrees, merging adjacent objects or suppressing a valid object. In this work, we assign an embedding vector to each pixel through a deep neural network. The network is trained to output embedding vectors of similar directions for pixels from the same object, while adjacent objects are orthogonal in the embedding space, which e ectively avoids the fusion of objects in a crowd. Our method yields state-of-the-art results even with a light-weighted backbone network on a cell segmentation (BBBC006 + DSB2018) and a leaf segmentation data set (CVPPP2017).
- Instance Segmentation of Nematode Cysts in Microscopic Images of Soil Samples
Long Chen, Martin Strauch, Matthias Daub, Marcus Jansen, Hans-Georg Luigs and Dorit Merhof
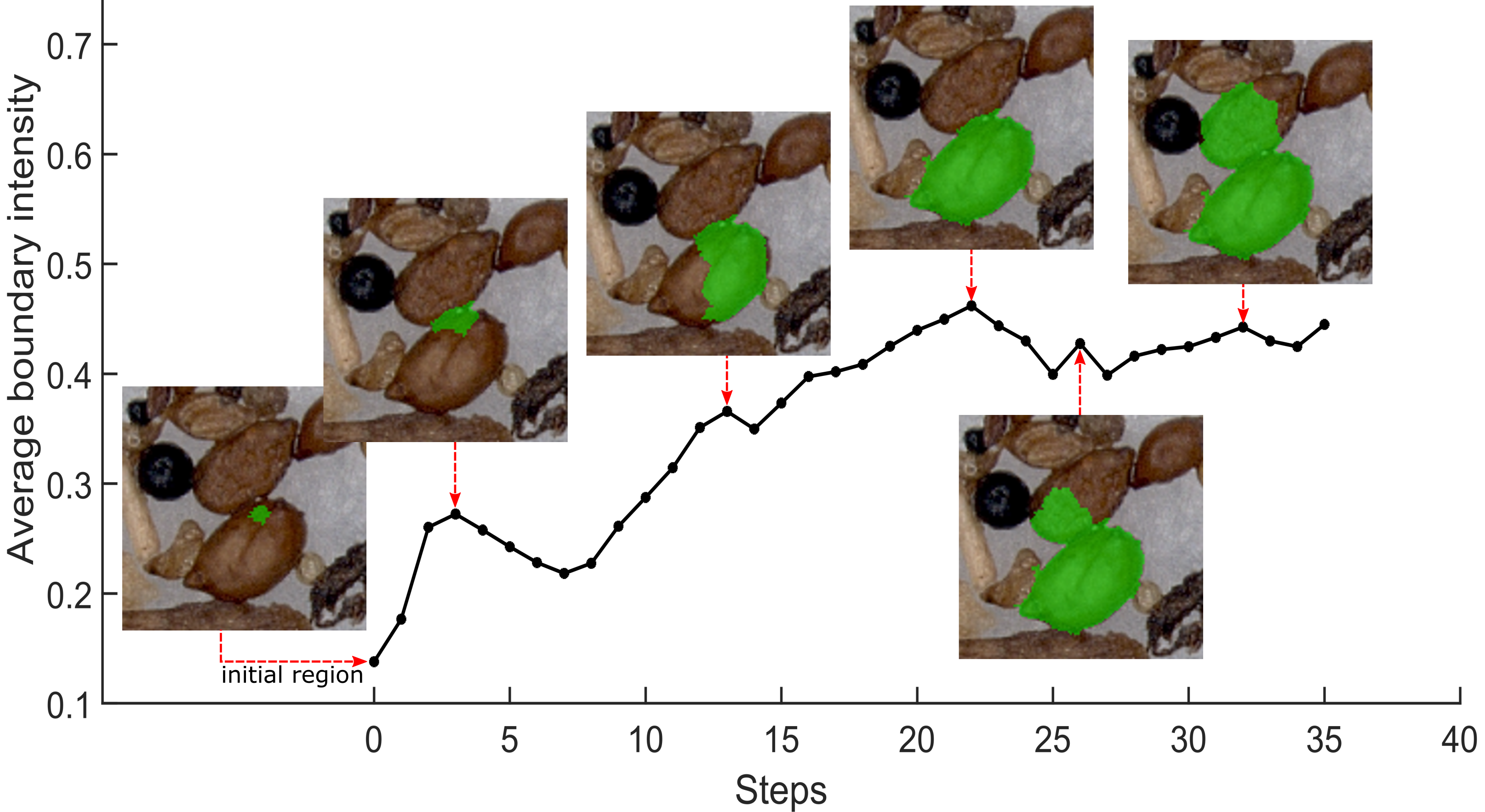
Abstract: Nematodes are plant parasites that cause damage to crops. In order to quantify nematode infestation based on soil samples, we propose an instance segmentation method that will serve as the basis of automatic quantitative analysis. We consider light microscopic images of cluttered object collections as they occur in realistic soil samples. We introduce an algorithm, LMBI (Local Maximum of Boundary Intensity) to propose instance segmentation candidates. In a second step, a SVM classifier separates the nematode cysts among the candidates from soil particles. On a data set of soil sample images, the LMBI detector achieves near-optimal recall with a limited number of candidate segmentations, and the combined detector/classifier achieves recall and precision of ~0:7. The pipeline only requires simple dot annotations and moderately sized training data, which enables quick annotating and training in laboratory applications.
- MixNet Multi-modality Mix Network for Brain Segmentation
Long Chen and Dorit Merhof
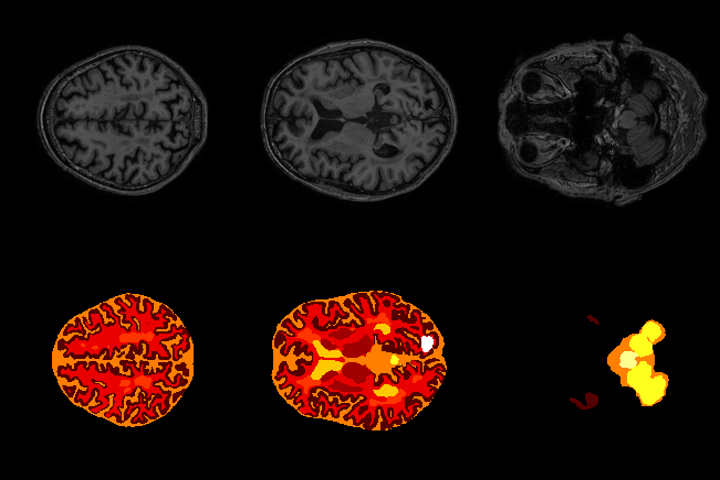
Abstract: Automated brain structure segmentation is important to many clinical quantitative analysis and diagnoses. In this work, we introduce MixNet, a 2D semantic-wise deep convolutional neural network to segment brain structure in multi-modality MRI images. The network is composed of our modi ed deep residual learning units. In the unit, we replace the traditional convolution layer with the dilated convolutional layer, which avoids the use of pooling layers and deconvolutional layers, reducing the number of network parameters. Final predictions are made by aggregating information from multiple scales and modalities. A pyramid pooling module is used to capture spatial information of the anatomical structures at the output end. In addition, we test three architectures (MixNetv1, MixNetv2 and MixNetv3) which fuse the modalities differently to see the e ect on the results. Our network achieves the state-of-the-art performance. MixNetv2 was submitted to the MRBrainS challenge at MICCAI 2018 and won the 3rd place in the 3-label task. On the MRBrainS2018 dataset, which includes subjects with a variety of pathologies, the overall DSC (Dice Coeffcient) of 84.7% (gray matter), 87.3% (white matter) and 83.4% (cerebrospinal fluid) were obtained with only 7 subjects as training data.